The Omicron variant may struggle to sustain community spread due to large vaccine rollouts and the Australian population’s improved antibody responses, write Stuart Turville, Anupriya Aggarwal, Anouschka Akerman and Vanessa Milogiannakis …
THE arrival of Omicron BA.1 in late 2021 started Australia’s journey into observed high case rates of community transmission. Closely following BA.1 was Omicron BA.2, a genetically distinct variant not derived from BA.1, which was co-circulating and clearly fitter. From this point on, all variants (except for the Delta–Omicron recombinants) were derived from a BA.2 parent. The first globally dominant variant derived from BA.2 progeny was BA.5 and in mid 2022 this drove many waves globally. Although the changes in BA.2 to BA.5 were not extensive and were confined primarily to the receptor-binding domain of the spike, they were in the right place to better navigate antibody responses and sustain a fitness advantage over BA.2 to finally supplant the latter parent. Yet towards the end of 2022, BA.5 was slowly fading, and unlike previous waves, several variants emerged in co-circulation. This period of the pandemic and variants was defined by what we referred to as a convergence. Many variants appeared with distinct evolutionary paths, but they all shared the accumulation of a similar and sometimes identical cluster of changes within the spike glycoprotein. Changes included R346T, L452R/M, F486V/I/P, N460K, K444R/T and F490S; all within the vicinity of the receptor-binding domain of the spike where the virus engages the primary receptor ACE2, but also where many of our neutralising antibodies bind and block/neutralise the virus. In following the antibody responses at the population level, we observed the pressure faced by the virus over time. Large vaccine booster rollouts, large Omicron case waves, and maturation of antibody responses over time were creating greater pressure on the virus to remain competitive in sustaining community spread. The convergence of many variants with similar spike glycoprotein changes was the result of their need to navigate this maturing response to remain competitive.
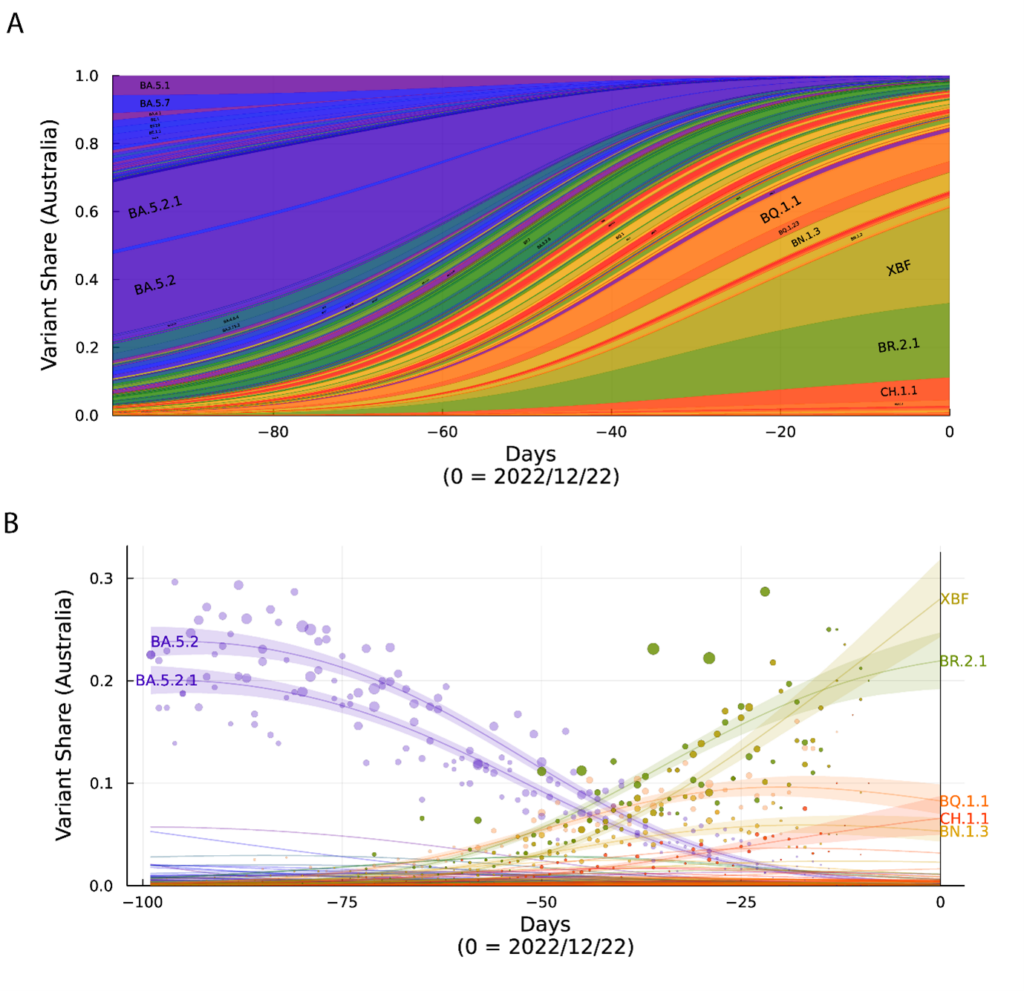
A. Surveillance summary of Australia at 22/12/22 based on genomic data via GISAID. Each vertical slice depicts the posterior mean variant frequency estimate from a hierarchical Bayesian multinomial logistic model of variant competition. Note the two dominant variants BR.2.1 and the recombinant XBF which are dominating in the states of NSW and Victoria, respectively. B. Variant proportions with associated Bayesian Credible Intervals for A.
Whilst the world experienced many genetically distinct variants in co-circulation, was Australia and other[GP1] countries experiencing the same or a unique set of co-circulating variant clusters? As the Northern Hemisphere is concerned with newly emerging variants such as the antibody evasive BQ.1.1 variant and now the recombinant XBB.1.5, Australia experienced enrichment of two specific variants, BR.2.1 and the recombinant XBF (Figure). Although there has been significant traction in the media of the variant XBB.1.5 that initially spread in North America, its appearance in Australia has only led to date of 16 confirmed detections nationwide over the Christmas period of December and January. While it may expand its footprint in Australia over time, it is a reminder that a variant that is competitive in one jurisdiction may not be competitive in another. Even if a variant has a competitive advantage, it would need to be of significant magnitude (like Alpha to Delta or more recently Delta to Omicron BA.1 for instance) in order to rapidly globally dominate in what can be described now as global immunological mix. And this mix is indeed different. In North America, early vaccine rollouts resulted in a greater time periods between booster shots. In Australia, however, the Delta wave increased vaccine uptake. Vaccine dosing and subsequent booster shots were compressed to a shorter time period and in theory based on past observations of vaccine dosing intervals this may have given less time for our immune systems to mature antibodies responses. The subsequent rollout of different booster shots may further diversify jurisdictional antibody responses, where BA.5 and BA.1 bivalents are being used in contrast to the original vaccine formulations or completely different formulations altogether (eg, Coronavac). Combine this with various high case numbers of BA.1, BA.2 and BA.5 Omicron waves and the community responses may gain further diversification in the context of potency and breadth in responding to each emerging variant. While the immunological landscape may shape which variant is enriched in each country, there are many other variables that can influence their distribution. Indeed, a perfect working example of this would be the variants rapidly spreading throughout China and Japan. In both countries, it is the early BA.5 sub-lineages that Australia and other countries had experienced in mid‑2022 that have primarily spread there, with lineages like BQ.1.1, BR.2.1, XBF and XBB.1.5 constituting a very small footprint. Yet if we compare the two countries with respect to their vaccine type, booster uptake and case waves over the pandemic, they are indeed different. China primarily used the Coronavac vaccine (inactivated virus) combined with limited reported cases throughout the majority of 2022; Japan primarily used mRNA vaccines (BNT162b2 and mRNA-1273 ) and experienced large case waves commencing in early 2022 with the arrival of BA.1 and BA.2. So at present, with many co-circulating variants with similar fitness, it will be interesting in 2023 to observe if countries like those listed above continue to sustain their own unique variant mix. This mix will indeed impact monoclonal antibody therapeutics and may also influence the need to increase booster uptake. The unique variant make-up in each country will need to be considered to inform policy settings and subsequent responses regarding vaccine booster doses and available therapeutics that still retain activity.
The situation is of course dynamic and we do not yet know if complacency in booster uptake over time will take place, and how this will impact this trajectory. Although the mobilisation of new bivalent vaccines is promising, we need to recognise vaccine inequality globally has the potential to drive variants to enrich differentially in each country. While it is important to continue variant surveillance, novel solutions to track the status of antibodies at the population level in Australia and other countries would be a pragmatic means to monitor the jurisdictional risk to emerging variants over time. Furthermore, if new vaccine formulations are shown to generate broader responses to future proof against emerging variants, fast-tracked distribution for global access would be a far superior means to increase pressure on virus spread on many fronts. Understanding and breaking through the bottleneck of global distribution needs refocused attention, as the global spread of the virus by no means is restricted to the degree it was earlier in the pandemic.
Although BR.2.1 and XBF dominated our variant make-up in late 2022, accompanying them was a very large list of other variants that appeared but never gained traction. It is important to keep track of variants that start to compete in other countries, but there are two important lessons that we learnt from the variant mix in Australia in late 2022. First, we need to move beyond the conversation of talking about each variant at a singular level. There are literally hundreds of Omicron types that can be distinguished by their genetic footprint and even now in Australia we have two co-dominating in our variant mix. While some may be more competitive (evade antibodies better or have a fitness edge in transmission), they are all variations on the same theme and pragmatically may indeed be better considered as various Omicron subgroups. Over time, many converge to share the same changes but are from different viral “parents”. How they enter our body has not diverged significantly to the point where one can be differentiated by targeting a different cell or tissue (ie, a significant shift in viral tropism). Secondly and importantly, we need to continue our surveillance of our own variants in Australia at both the genetic and phenotypic level. This is presently led and performed by many excellent teams at the state level. However, continued support for these diagnostic and surveillance teams is needed. We will also need to consider current changes to COVID-19 testing policies and how this will influence genotypic and phenotypic surveillance, as at present, a PCR based test is the primary means that surveillance teams can gain access to samples for genomics and viral isolations. Furthermore, continued support is needed in bridging the diagnostic-research teams. The pandemic has brought together the power and expertise of many teams, which can continue to contribute at many levels and provide evidence to guide policy in managing COVID-19.
To conclude, while the variant mix in Australia has been unique and diverse in late 2022, it fortunately has not driven case waves like those we experienced with Omicron BA.1 in early 2021. It is important to be vigilant and continue to carry out genotypic and phenotypic surveillance of emerging variants, but we must also recognise that the immune landscape of our population has matured. This immune legacy is the consequence of many rounds of high vaccine uptake and large case waves, and through testing of many people who generously donated blood in vaccine and infection cohort studies, we can recognise and respond to many emerging variants in our present mix. Yet as we have highlighted above, this situation is dynamic. The longevity of the immune legacy obviously will take time to quantify. As the conversation about the virus leaves the front pages of our newspapers, where will the conversation lead? Will complacency and vaccine inequity, for instance, result in geographic pockets where the virus can readily survive? How many changes does the virus have that can sustain a viral phenotype that is fit in transmission but also able to navigate our antibody and other protective layers of immune response? Many of these answers are as yet unknown, but as we have experienced with other viruses, control through vaccination and/or therapeutics is the key to controlling viral load at many levels.
Associate Professor Stuart Turville leads virology and containment activities at the Kirby Institute, UNSW.
Dr Anupriya Aggarwal is a Research Fellow at the Kirby Institute and co-leads the laboratory with Associate Professor Stuart Turville and has engineered many assay platforms to aid in rapidly characterising emerging variants.
Dr Anouschka Akerman is an early career researcher leading many COVID-19 related projects at the Kirby Institute.
Ms Vanessa Milogiannakis: Is a research associate and PhD student researching the development of novel assay platforms to study emerging viruses.
The statements or opinions expressed in this article reflect the views of the authors and do not necessarily represent the official policy of the AMA, the MJA or InSight+ unless so stated.
Subscribe to the free InSight+ weekly newsletter here. It is available to all readers, not just registered medical practitioners.
If you would like to submit an article for consideration, send a Word version to mjainsight-editor@ampco.com.au.

 more_vert
more_vert