FOR anyone finding themselves on an unknown street these days, directions are just a few taps away on a mobile phone map.
By stitching together satellite imagery, aerial photography, and street maps, programs like Google Maps are something most people use every day.
Scientists and clinicians have been seeking the same kind of resource for the journey that cancer cells take throughout disease.
To grow and spread, cancer cells take a number of genetic steps to overcome the normal cell controls that allow them to enhance their growth rate, move into nearby tissues and evade our immune system.
So, instead of satellite images, researchers have submitted human tumour samples from all over the world, piecing together their genetic information into the recently published Global Cancer Atlas.
The atlas is composed of the entire cancer genomes from more than 2,600 patients suffering from 38 different tumour types. The cancer types range from common forms like colorectal and breast cancers, to rare cancer types including pancreatic and brain cancers.
The 1,300 scientists and clinicians behind it belong to the Pan-Cancer Analysis of Whole Genomes Project (PCAWG), which gets its name from the 37 countries that have worked together for a decade to produce the map, now published in the journal Nature.
Just as the number of streets in the world is finite, so too are the genetic steps that result in cancer, despite being extremely complicated, says Professor Sean Grimmond who has led the Australian team of the International Cancer Genome Consortium (ICGC).
“Every cancer is a result of genetic scars, which is damage that occurs to DNA over time,” says Professor Grimmond, from the University of Melbourne Centre for Cancer Research.
“By combining sequencing of the whole cancer genome with a suite of analysis tools, we can characterise every genetic change found in a cancer,” he says.
“This includes all of the processes that have generated those mutations, and even the order of key events during a cancer’s life history, the kind of information that a good route-planning resource will tell you,” he says.
Scientists had already analysed cancer genetics for all of the genes that produce proteins, known as the exome or coding regions. However, 99 per cent of DNA – our genetic blueprint – does not code for proteins, but still includes important regions like those responsible for switching genes on and off.
“The information we previously had was similar to knowing all of the streets in the coasts of the world, but nothing in between, so the Global Cancer Atlas was undertaken to look at changes in every part of the DNA,” Professor Grimmond says.
To begin the project, every cancer tumour was described by a pathologist including its tissue of origin and cancer stage. The cell’s entire DNA sequence was determined and then uploaded into a cloud data-sharing project for analysis in order to the compare the tumours.
“What the Cancer Genome Atlas tells us is that for every cancer there are approximately five genetic scars,” he says. “The combination of genetic scars responsible for each cancer type is complex and involves dozens of different genes for each cancer type,” Professor Grimmond says.
The work helps explain a long-standing medical challenge, where two patients can present with the same type of cancer, but they would respond very differently to the same treatment.
The head-to-head comparison of each cancer type in the atlas also identified surprising commonalities in driving mutation pattern between individuals suffering from different tumours which grow in different sites.
“This is particularly exciting as it provides a rationale for future testing of drugs known to be successful in one cancer type in patients with very different tumours that bear the same driver mutation profile.”
The types of genetic scars identified by the Global Atlas range from single changes in a DNA letter to deletions, flips or rearrangements of chromosomes that contain thousands of pieces of genetic information.
Some of the most surprising results were finding the sheer number of genetic changes, what we call mutation signatures. We had previously identified 21 main types of signatures in pancreatic cancer for example, but work on the Cancer Atlas has revealed 90.
These changes can occur years, even decades before the cancer occurs and each type of change of mutation has a distinct signature. This uniqueness is a huge advantage when a patient presents with a cancer that has moved or metastasized.
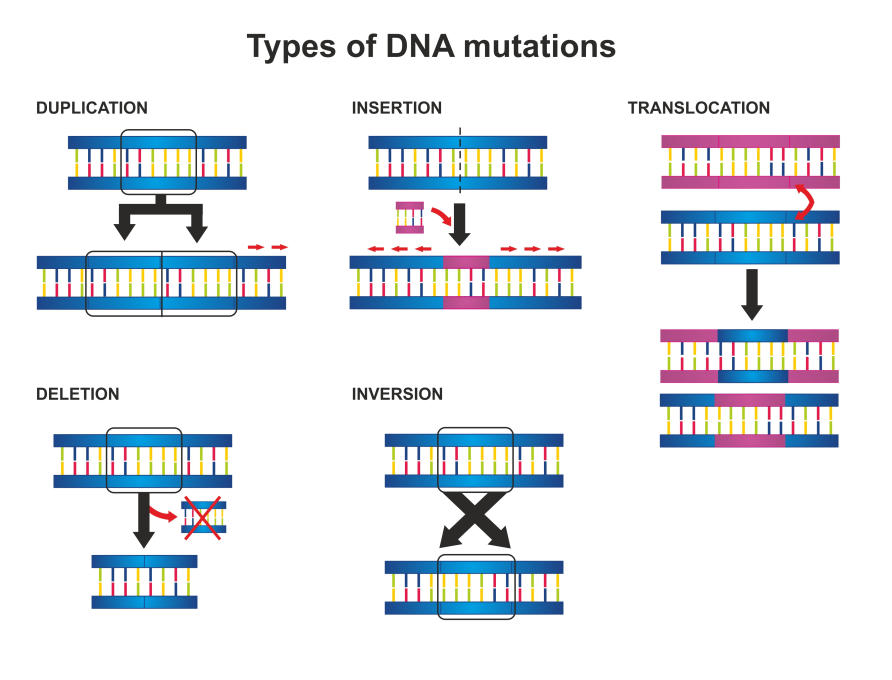
“If we don’t understand where a cancer comes from, we can’t even rely on traditional clinical approaches to treatment,” Professor Grimmond says.
This unprecedented level of data crowd-sourcing means researchers, clinicians and industry can now work together, anywhere across the globe.
“We have made all of the genetic information and analysis tools open access, for the fastest collaboration and development of new diagnostics and treatments,” Professor Grimmond says.
“Having a harmonised dataset enables international researchers to learn from one cancer treatment and apply those findings to another using a cloud computing portal.”
The next step is to add even more information to the atlas, the team aim to add ten times as many patient samples, analysing each patient at a time to understand how common each pattern of genetic damage is.
“Although the genome of each patient’s cancer is unique, there are a finite set of recurring patterns of genetic damage, so with large enough studies we can identify all of them,” Professor Grimmond says.
“Our goal is for the Atlas be used as a reference to detect cancers earlier, to provide a personalised journey for the diagnosis and treatment of every patient.”
The Australian research groups involved include the University of Melbourne, Peter MacCallum Cancer Centre, QIMR Berghofer, Garvan Institute, University of Queensland and the Melanoma Institute of Australia.
This article was first published on Pursuit. Read the original article.
Dr Nerissa Hannink is a Senior Media Advisor at the School of Science, Veterinary and Agricultural Sciences at Melbourne University.

 more_vert
more_vert